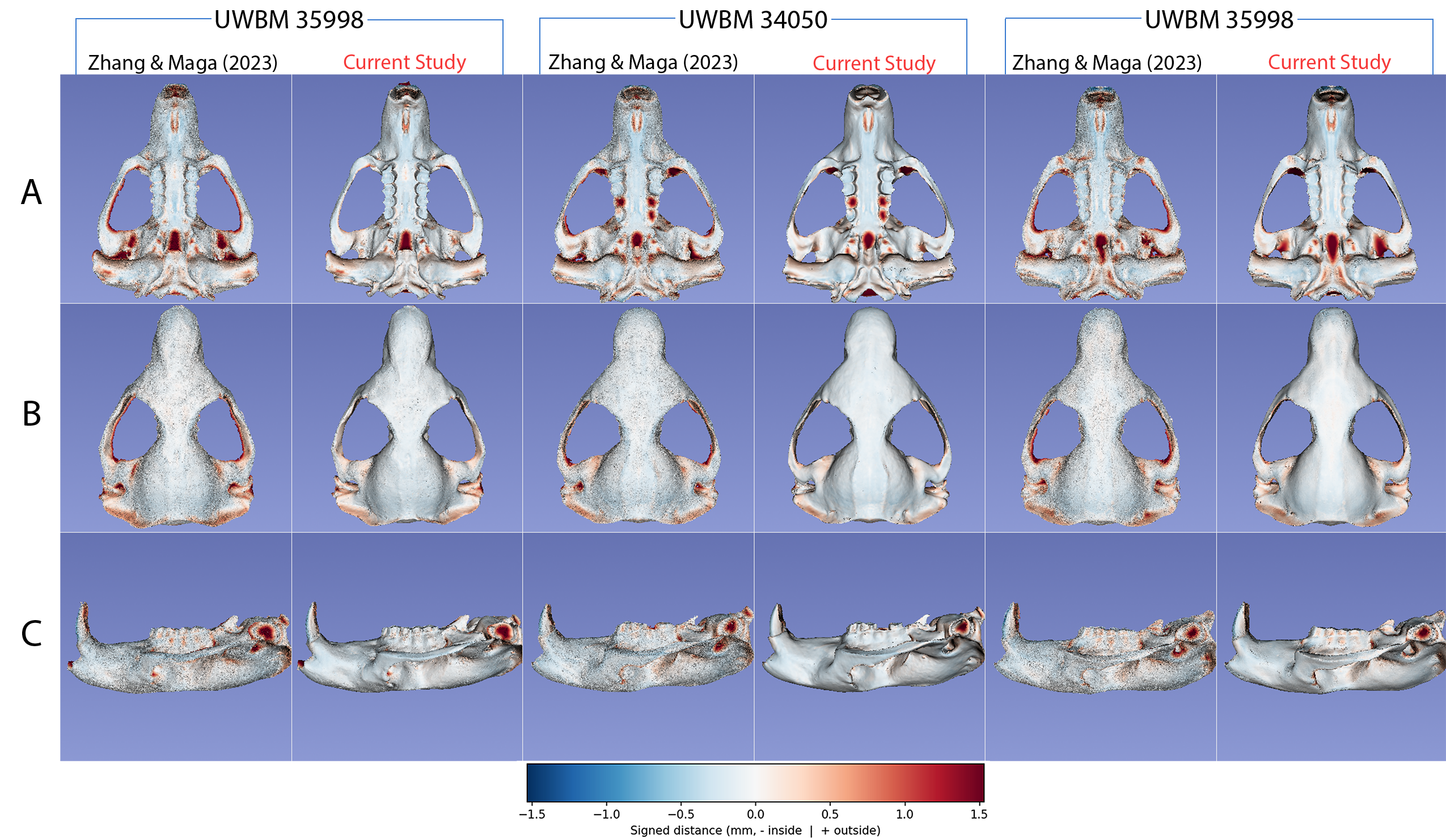
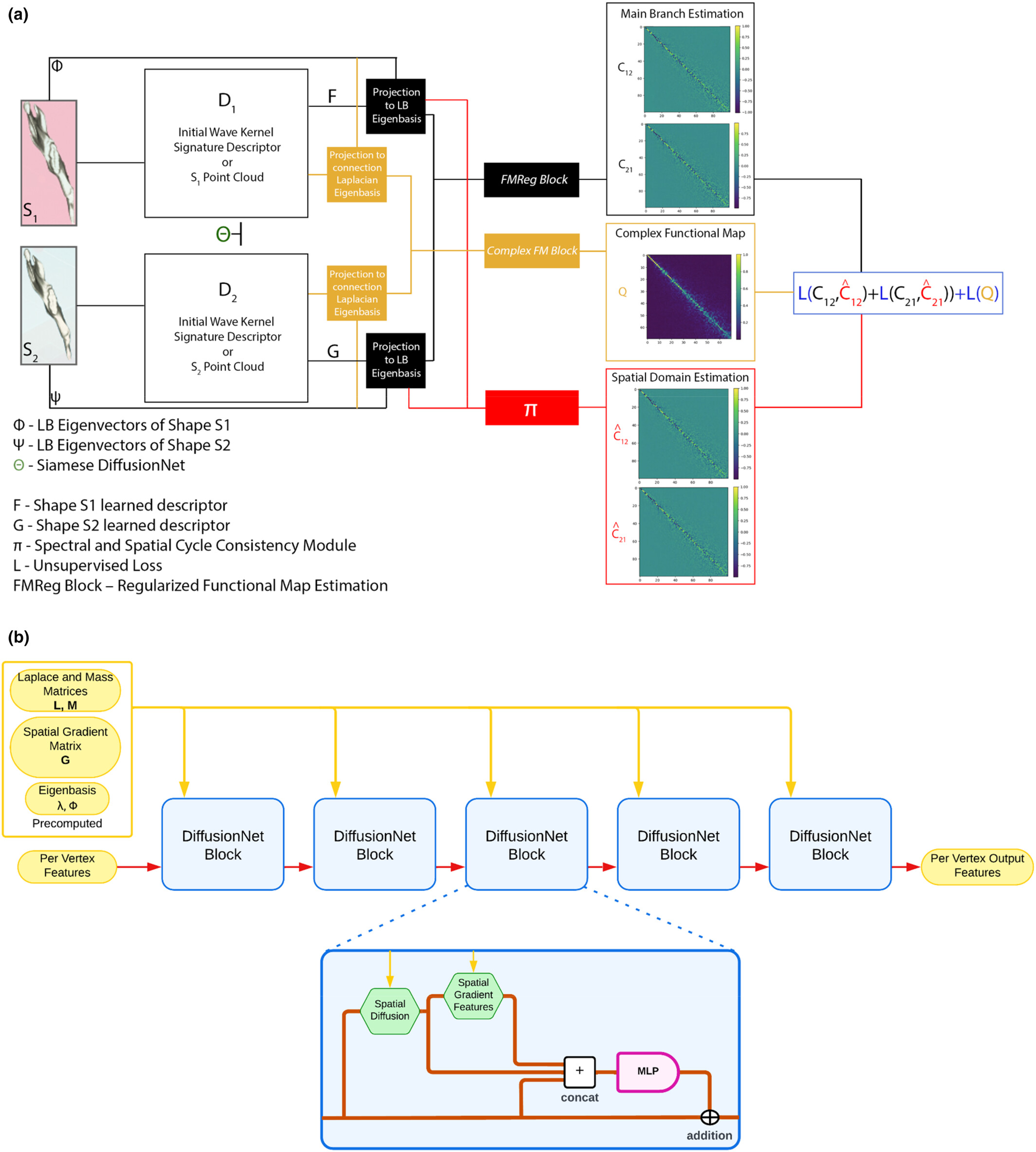
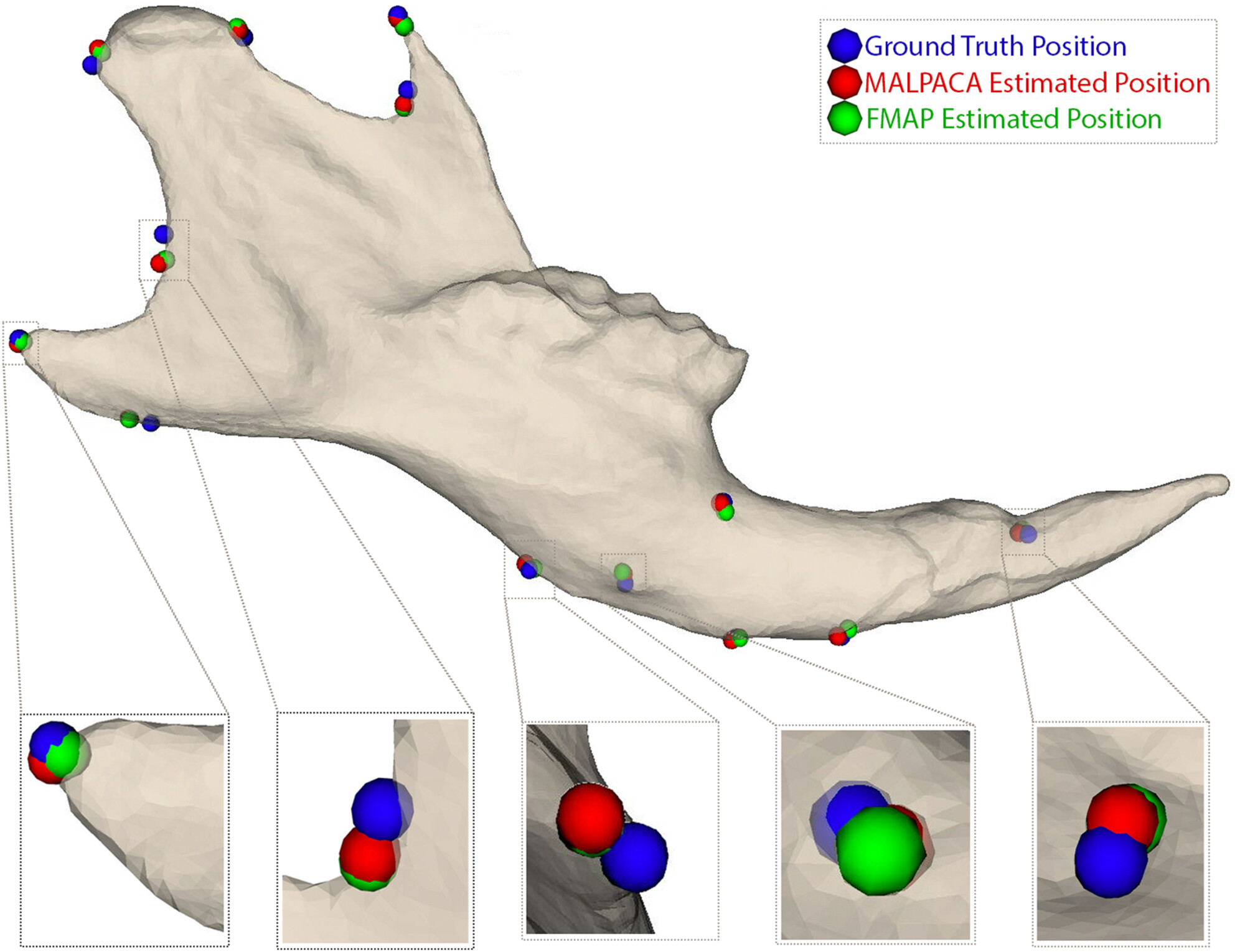
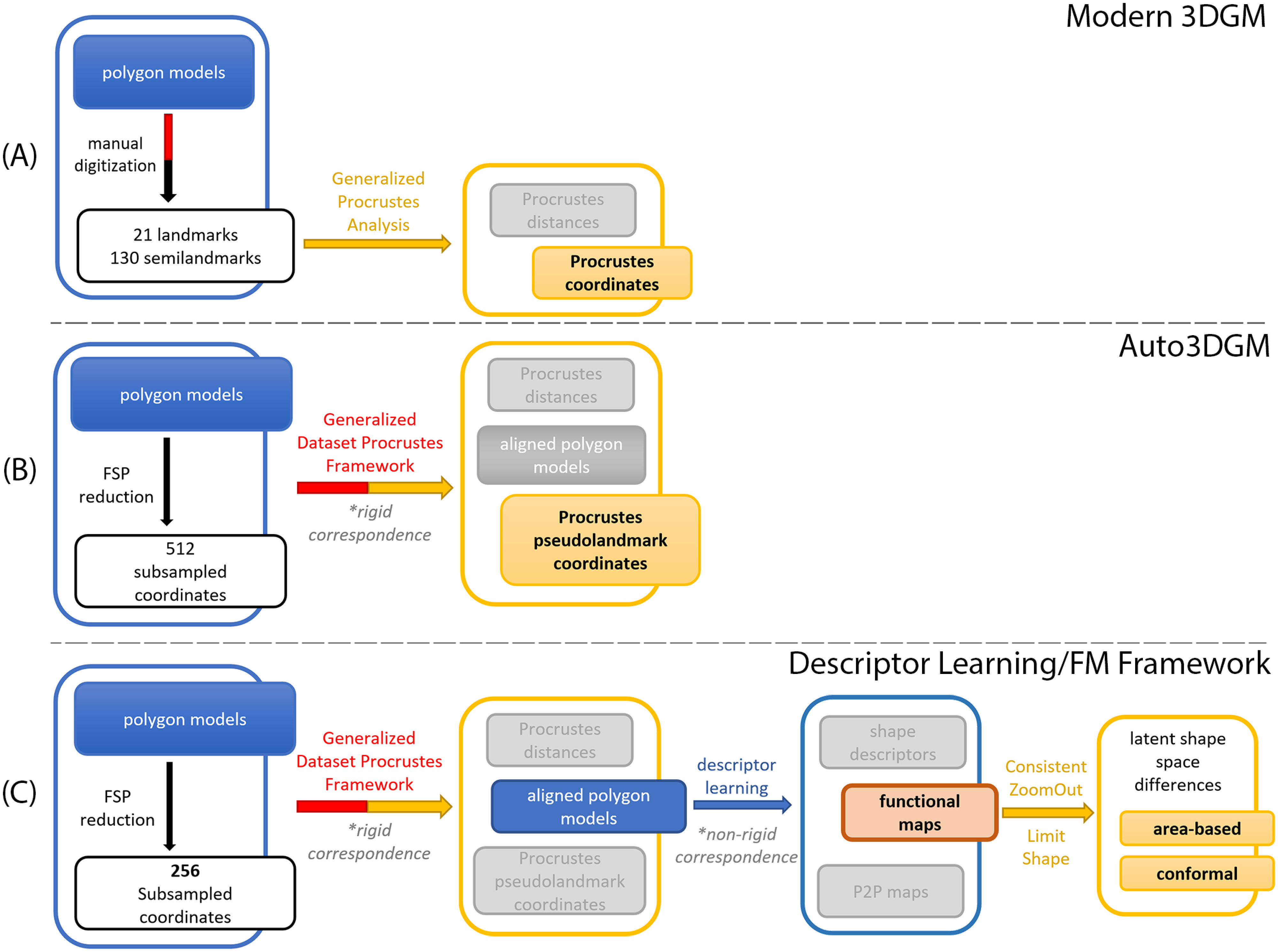
Trustworthy Exencephaly Detection with Focal‑Loss Transformers
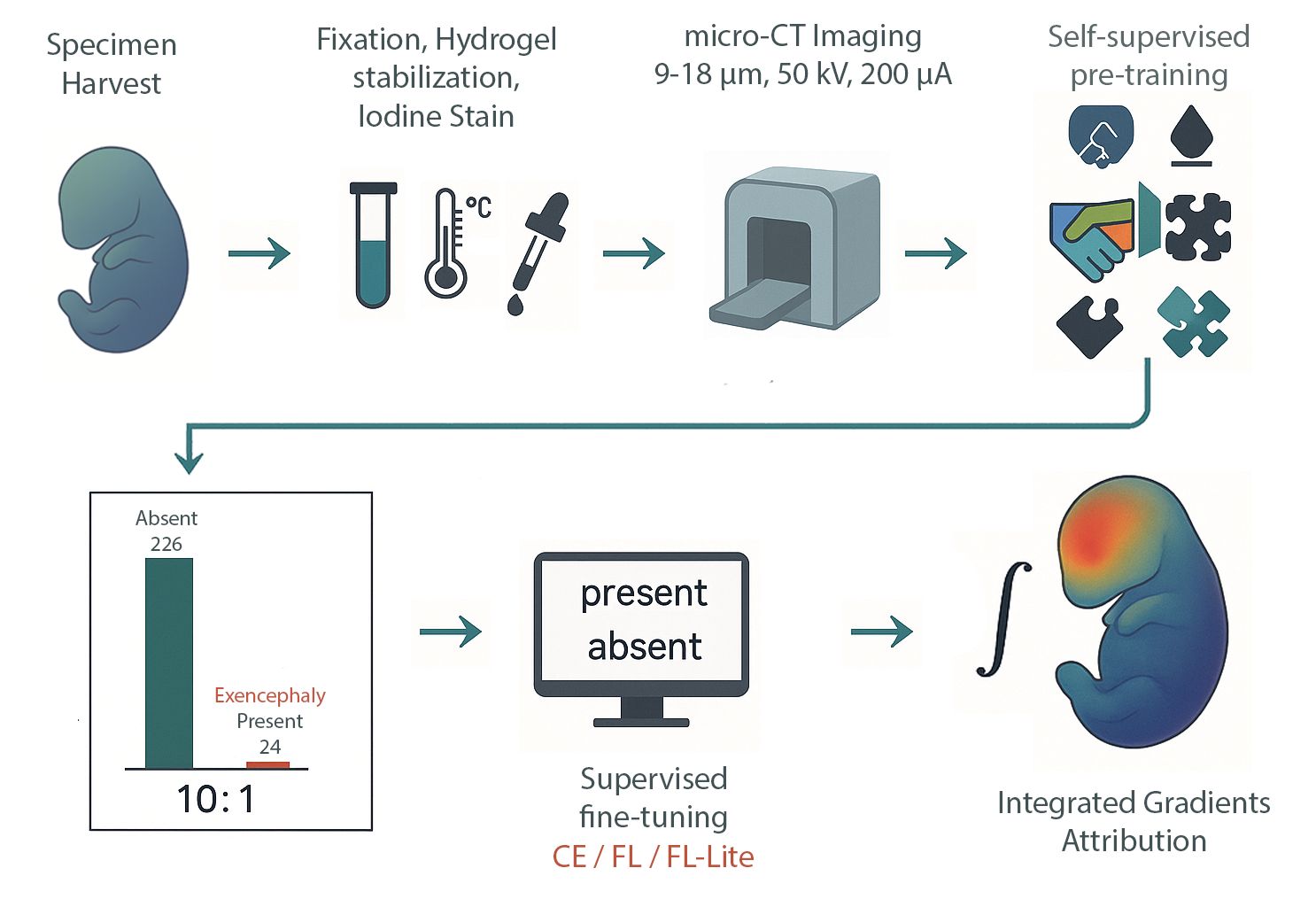
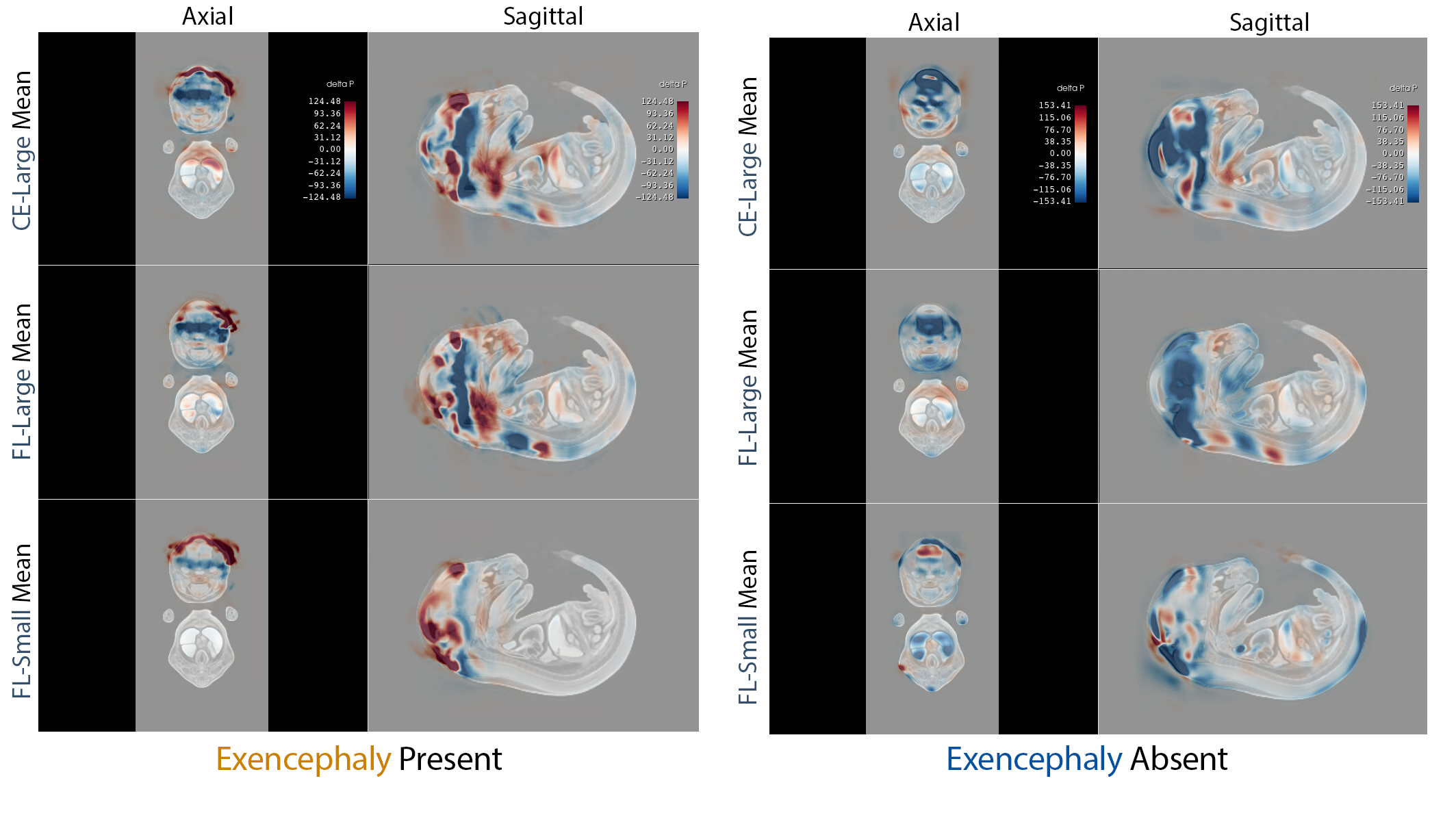
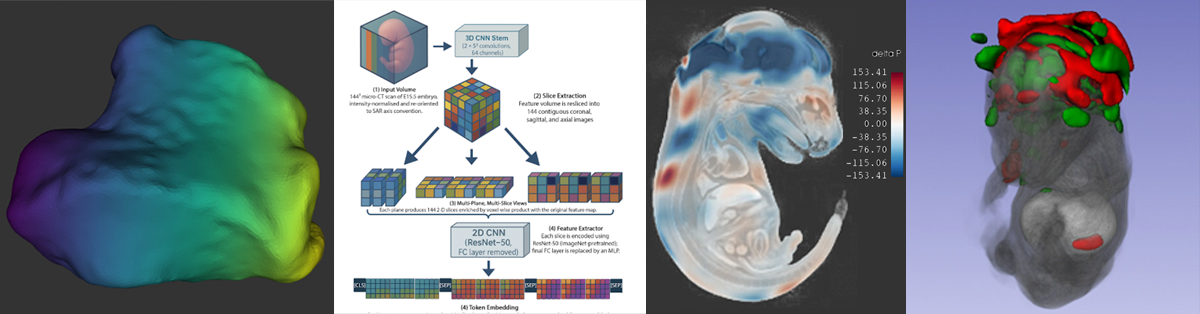
IMPC/KOMP‑style whole‑embryo iodine‑contrast micro‑CT (diceCT) screens search for lethal malformations across knockout lines—but positives are rare (~10%), a recipe for brittle classifiers and noisy saliency. We built an imbalance‑aware transformer that pairs focal loss with capacity control and seed ensembling, and we evaluated explanation quality—not just ROC. On 253 E15.5 volumes (24 exencephaly), all 15 models achieved 0.996 ± 0.002 accuracy; focal loss cut saliency entropy by up to 1.5 bits and doubled cross‑seed Dice, focusing attribution on the malformed cranial vault.
This urns attribution/saliency from impressionistic heatmaps into anatomy‑specific, reproducible evidence—without voxel labels—using only atlas registration and standard preprocessing.
Our approach delivers trustworthy, high‑throughput phenotyping for large mouse genetics programs, triaging defects with near‑perfect sensitivity and reproducible explanations. The recipe generalizes to other malformations and cohorts, offering a practical template for interpretable 3D classification under severe class imbalance.